KnotInFrame predicts -1 frameshift sites with simple pseudoknots. More complex knotted structures like triple crossing helices or kissing hairpins are excluded from calculation.
The prediction is based on a comparison between the minimal free energy (mfe) structure, calculated by an RNAfold like program and the mfe-structure, computed by a modified version of pknotsRG-mfe, called pknotsRG-frameshift developed by Jens Reeder and Robert Giegerich.
Short sequences up to 120 nucleotides can be predicted within 0.6sec. Genomes of 5MB can be compute in about 1h 37min.
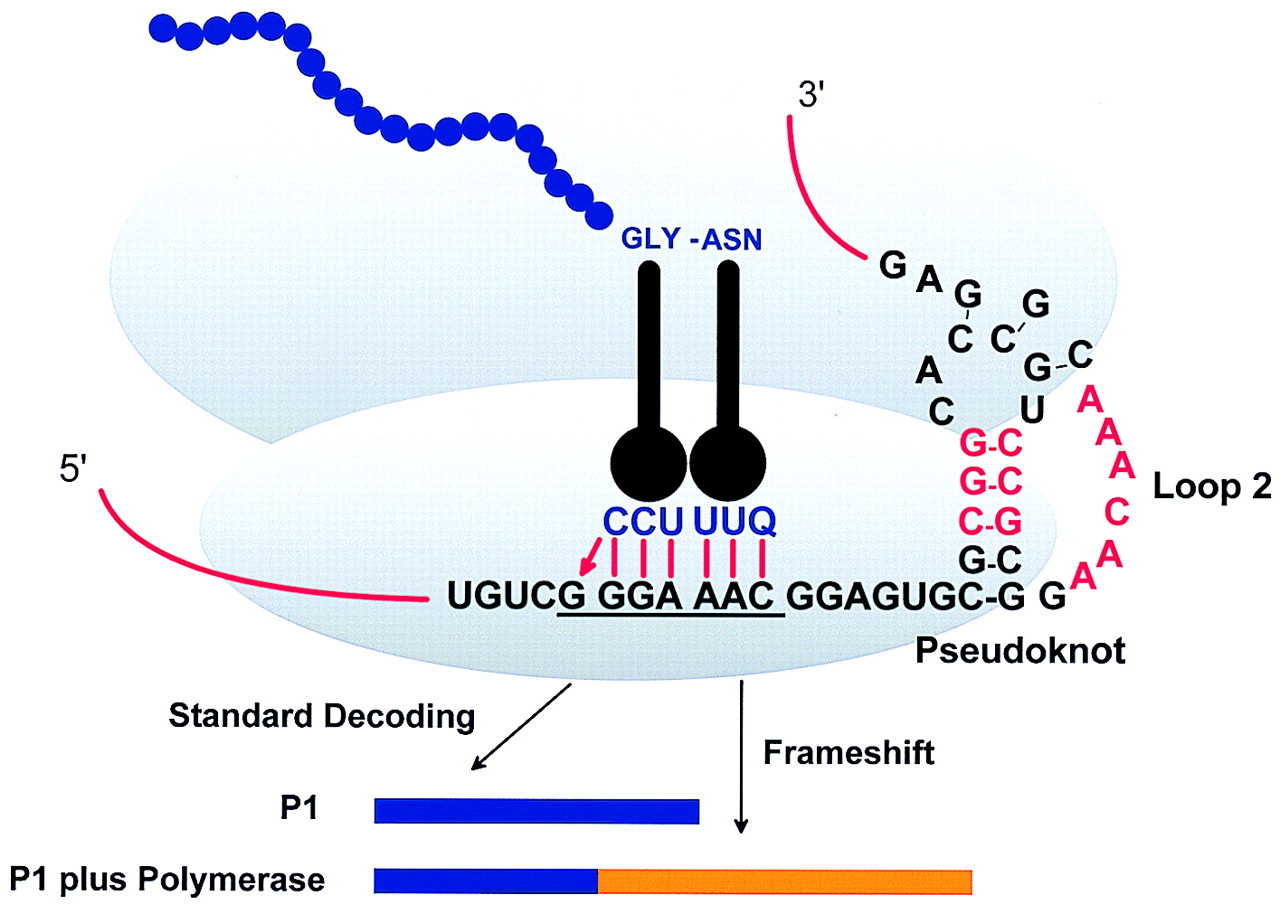
Figure 1: Model for BWYV -1 frameshift [
ala:atk:ges:1999].
Additional material on the
comparison with the yeast screen of [jac:bel:rak:din:2007]
