Pairwise alignment method function, provides most of the functionality of rnaforester command line tool. Offers two different example
for a local and global alignment.
Multiple alignment method function, provides most of the functionality of rnaforester command line tool. Offers one example.
Global, local and small-in-large alignment
Local similarity means finding the maximal similarity between
substructures of RNA secondary structures. If these substructures
are extended, the score decreases. This requires a scoring scheme
that balances positive and negative scoring contributions.
Otherwise, the similarity of the complete structures would always
achieve the maximum score. It is generally assumed that an
alignment of two empty structures scores zero. Local distance
makes no sense, as empty forests have always the lowest possible
distance of zero.
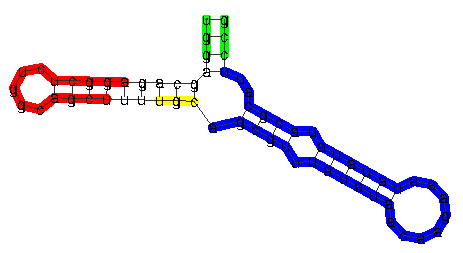
Substructures of RNA secondary structures in RNAforester are
contiguous and ``closed'' by hairpin loops. The blue region shows
a valid substructure. The green part of the structure is not
closed because the closing hairpin is missing. The red part is a
substructure that is not considered as a local structure for the
same reason. This is less obvious, since only theU,
which is a child of the root of this subtree, is not included. If
the top-levelP node would not be included in the red
substructure, this part would correspond to a closed subforest.
The yellow part does not correspond to a closed subforest since
the subtrees are not consecutive siblings.
Scoring models
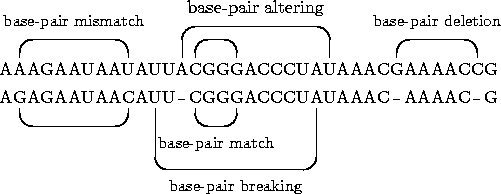 Structural edit operations of Jiang et al.'s general edit model
for RNA structures.
Structural edit operations of Jiang et al.'s general edit model
for RNA structures.
The sequence edit operationsbase match,base
mismatch andbase deletion are the same for
pairwise and multiple alignment. Abase pair breaking
means the deletion of a base-pair bond. Abase-pair
deletion is the composition of abase-pair breaking
and twobase deletions. Abase-pair altering is
treated likewise but there is only onebase-deletion
involved. The structural edit operationsbase pair
replacement and have a different effect for pairwise and
multiple alignment. In pairwise alignment mode, the pairing bases
are treated as a unit. In multiple alignment mode,base pair
replacement score means the score for matching any base-pair
plus the score for matching or mismatching the bases that pair.
Thus, it is not possible to construct a base-pair dependend
scoring for this model. The RIBOSUM scoring scheme are
empirically derived base-pair and single base substitution scores
that are available in pairwise alignment mode.
